Ph.D. Thesis
Title Rational Design of DNA Binding Molecules
Advisers Prof. Isao Saito
Thesis Committee Prof. S. Kitagawa and Prof. T. Imanaka, Department
of Synthetic Chemistry and Biological Chemistry, Kyoto University,
Japan.
Essay
The 21st century opened with the complete sequencing
of the human genome, providing useful tools, new subjects, and opportunities
for medical advance. Single nucleotide polymorphisms (SNPs), genetic
mutations of single nucleotide bases, are the most prevalent type
of DNA sequence variation found at a frequency of 0.5-10 per every
1000 base pairs in the human genome. SNPs are of great interest because
they are the mutations that cause the overwhelming majority of genetic
diseases and they can also serve as genetic markers. Recent research
has revealed a close relationship between the SNPs and drug metabolism,
spotlighting the SNPs as a potential indicator for drug action. There
have been increasing demands for high-throughput SNP detection.
SNPs are silent.Nucleotides of both mutant and wild-type
genes are completely intact with respect to chemical structure and
form a normal Watson-Crick base pair,making it very difficult to detect
SNPs without sequence information. However, when a wild-type gene
is mixed with a mutation gene, SNPs are emphasized as mismatched base
pairs in the resulting heteroduplex (the heteroduplex method developed
by Meyers et al.). This heteroduplex method opened up a way to directly
arrest the silent SNPs as mismatched base pairs, providing chemists
with a challenge to develop a strategy to detect mismatched base pairs
within a large amount of genome sequences. In my thesis work, we worked
to develop a strategy for rational design of small molecules that
bind to mismatched base pairs and, moreover, to apply this chemical
approach to a SNP scanning system.
Repair enzymes normally recognize mismatched DNA sites
in nature. Uracil-DNA glycosylase recognizes mismatched uracil in
DNA via a �nucleotide-flipping � mechanism, in which a uracil is flipped
out and trapped by the enzyme pocket and the resulting �hole � at
the flipped out uracil site is simultaneously occupied by a lysine
residue from the enzyme. The recognition system is very complicated
on the molecular level but can be represented as more general �push-pull
�mechanism. This realization inspired us to propose a conceptuall
new approach for DNA mismatch recognition.
Our strategy is quite simple and based on rational �push-pull�chemistry.We
hypothesized that the dimeric form of aromatic intercalating agents,
possessing hydrogen bonding groups fully complementary to mismatched
bases,can simultaneously recognize both mismatched nucleotide bases
within the base stacks of a DNA duplex (Figure 1).
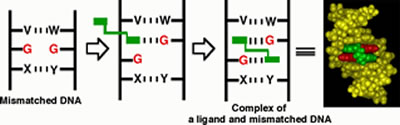
Figure
1. Molecular design of a ligand selectively binding to mismatched
base pair. Mismatched bases are colored red. Designed bis-intercalator
is colored green.
One of the mismatched bases is captured by one side
of the hydrogen-bonding intercalator and the resulting hydrogen-bonded
pair is stabilized within the p-stack of
DNA. The other half of the dimer fills the hole and binds stably to
the other mismatched base. It is very difficult to design a ligand
possessing a huge hydrophobic pocket to flip out the mismatched nucleotide
base, but in our strategy this difficulty is circumvented by utilizing
the DNA p-stack itself as an enzyme pocket
mimic. This small organic molecule (<500 Da) is a sophisticated mimic
of the naturally occurring enzyme (>10,000 Da).
First, we synthesized a small ligand called dimeric
naphthyridine (DNP), which is the dimeric form of a hydrogen bonding
intercalator fully complementary to guanine (G), and investigated
its binding to a G-G mismatch.G-G mismatches are one of the most stable
mismatched base pairs. Quantitative binding analysis showed that DNP
strongly binds to G-G mismatches with a very low dissociation constant
(53 nM) and selectively binds to G-G mismatches over other mismatched
and normal base pairs with more than 360-fold higher affinity.
To determine the mode of the binding of DNP to G-G mismatched
base pairs, we analyzed the 1D and 2D NMR spectra of the complex.The
2D-NOESY experiments clearly showed that each intercalator of DNP
simultaneously recognizes a guanine base in the G-G mismatch within
the DNA p-stack. Structure-activity and
energetic studies also clarified the mode of the binding, revealing
that cooperative capture of both G-bases is indispensable for mismatch
recognition. Based on these observations, we concluded that our conceptually
new strategy is very appropriate for the design of mismatch-binding
molecules.
Finally, we applied these results to the development
of a mismatch-detecting sensor that can be applicable to high-throughput
SNP scanning. We covalently immobilized the designed G-G mismatch
binding ligand, DNP, on a gold-chip surface. When mismatched DNA is
captured by immobilized DNP, the mass change induces a change of surface
plasmon resonance (SPR) on the gold surface, because SPR is directly
related to the total molecular weight of biomolecules on gold surface.
The accuracy of this system was demonstrated by a marked SPR response
obtained only for synthetic DNA containing a G-G mismatch while other
mismatches produced only a weak response. Finally, with this system,
we succeeded in detecting mismatched base pairs in heteroduplexes
produced from real biological samples, specifically a 652 base pair
PCR product of the human HSP70-2 gene. This SPR based method is suitable
for high-throughput scanning of SNPs, because it is simple, inexpensive,
and reproducible. This sensor can be used repeatedly without any special
handling, each run of the assay finishes within 30 min, and this assay
does not require any labeling of the DNAs.
In summary, my thesis work provided a new strategy to
design mismatch-binding ligands that bind with high affinity and selectivity.
The chemistry described here provides useful information for the development
of related ligands that bind to the other mismatches in DNA. The SPR
sensor-chip that makes use of this chemical strategy can be a promising
system for SNP scanning in the future, post genome era.
<Download full-text essay,
pdf file-105KB>

